Hi
My first post !
Context
I’m trying to make a mosaic with Mosaic Application and harmonization method to ‘band’.
Configuration setup
My system: ubuntu 20.04.1
Version of the OTB: 7.1
I installed the OTB with: don’t know…
Python version: 3.8.10
i’m using also pytob version: 1.4.0
Description of my issue
I’m doing a mosaic with three tiles of Sentinel-2. It’s not with raw bands but with a DHI (dynamic habitat index that is an annual synthesis of radiometric indices) of NDVI.
-
The mosaic with S2 raw bands and parameter
'harmo.method': 'band'is correct -
The mosaic with DHIs and and paramter
'harmo.method': 'band', the mosaic is strange and stats are :
MAX: 1056, MEAN: 223, MIN: -6844
Stats of the DHI used are:
tile T32TLR : MAX: 999, MEAN: 688, MIN: -493
tile T31TGL : MAX: 999, MEAN: 798, MIN: -998
tile T31TGK : MAX: 999, MEAN: 727, MIN: -994
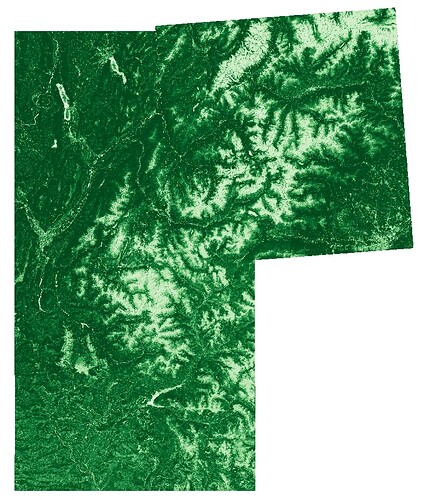
Here is a capture of the three independent DHIs:
And the mosaic:
The gains seems not to be normal but I don’t understand why:
Gains : 0.625725 0.292308 0
For another combination of DHIs the gains are different:
Gains : 0 0 0.295846
- The mosaic with DHIs and paramter
'harmo.method': 'none', the mosaic is correct but with no harmonisation… and at the connection between tiles there is no data values.
Here part of my script:
dhi_mosaic = pyotb.Mosaic({'il': tile_list,
'comp.feather': 'none',
'harmo.method': 'band',
'harmo.cost': 'rmse',
'interpolator' : 'bco',
'output.spacingx' : 100,
'output.spacingy' : 100,
'nodata': -10000
})
filename_extension = "?&streaming:type=tiled&streaming:sizemode=height&streaming:sizevalue=512"
dhi_mosaic.write(dhi_mosaic_path, pixel_type='int16', filename_extension=filename_extension)
Is someone can help me ?
Thanks !